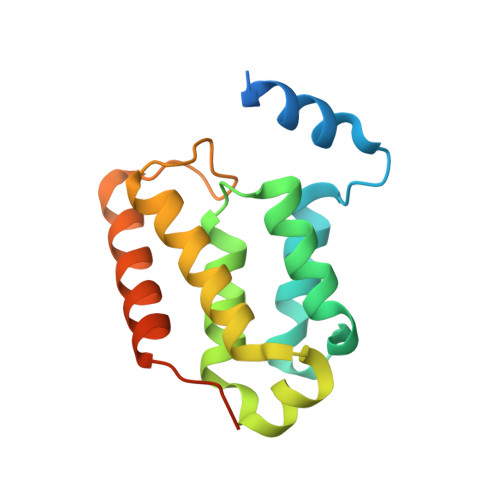
Crystal structure of the Leishmania major MIX protein: A scaffold protein that mediates protein-protein interactions.
Gorman, M.A., Uboldi, A.D., Walsh, P.J., Tan, K.S., Hansen, G., Huyton, T., Ji, H., Curtis, J., Kedzierski, L., Papenfuss, A.T., Dogovski, C., Perugini, M.A., Simpson, R.J., Handman, E., Parker, M.W.(2011) Protein Sci 20: 1060-1068
- PubMed: 21465610
- DOI: https://doi.org/10.1002/pro.631
- Primary Citation of Related Structures:
3HA4 - PubMed Abstract:
Infection by Leishmania and Trypanosoma causes severe disease and can be fatal. The reduced effectiveness of current treatments is largely due to drug resistance, hence the urgent need to develop new drugs, preferably against novel targets. We have recently identified a mitochondrial membrane-anchored protein, designated MIX, which occurs exclusively in these parasites and is essential for virulence. We have determined the crystal structure of Leishmania major MIX to a resolution of 2.4 Å. MIX forms an all α-helical fold comprising seven α-helices that fold into a single domain. The distribution of helices is similar to a number of scaffold proteins, namely HEAT repeats, 14-3-3, and tetratricopeptide repeat proteins, suggesting that MIX mediates protein-protein interactions. Accordingly, using copurification and mass spectroscopy we were able to identify several proteins that may interact with MIX in vivo. Being parasite specific, MIX is a promising new drug target and, thus, the structure and potential interacting partners provide a basis for structure-guided drug discovery.
Organizational Affiliation:
Biota Structural Biology Laboratory, St. Vincent's Institute of Medical Research, Victoria 3065, Australia.